References workflow¶
This workflow is not normally run on its own. Rather, it is intended to be include:-ed into other workflows that depend on reference fastas, indexes, and annotations. That way, rules in this references workflow will only be run for those files asked for in the parent workflow.
It is still possible to run this workflow on its own. In that case, it will build all of the references and indexes specified in the config. This can be helpful when setting up the workflows for the first time on a new machine.
In all cases, it depends on the references section being in
config/config.yaml. See References config for details on
configuring.
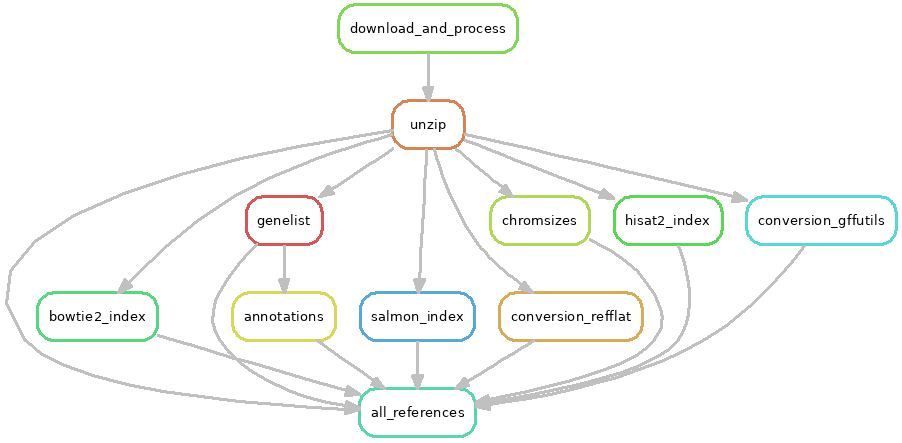
A dictionary of references¶
When run on its own, the references workflow in workflows/references/Snakefile
builds all references specified in the config file. This is typically done
only when initially setting up a system that will run workflows on many
different references.
Most of the time, this workflow is included into the other workflows with the
include: directive. This way, any reference files that are needed by, say,
the RNA-seq workflow will be created automatically.
The format of the config YAML is designed to be convenient to edit and
maintain. It can be awkward to use within a Snakefile though, so for
convenience it is converted into an easier-to-access dictionary in
the c config object in each workflow, accessible as c.refdict.
If we have the following references section defined in our config file (see Configuration for more):
references:
dm6:
r6-11:
fasta:
url: "https://url/to/dm6.fasta"
indexes:
- bowtie2
- hisat2
gtf:
url: "https://url/to/gm6.gtf"
conversions:
- refflat
r6-11_transcriptome:
fasta:
url: "https://url/to/transcriptome.fa"
indexes:
- salmon
then it will be converted to this simplified version where values are filenames:
{
'dm6': {
'r6-11': {
'fasta': '/data/dm6/r6-11/fasta/dm6_r6-11.fasta',
'refflat': '/data/dm6/r6-11/gtf/dm6_r6-11.refflat',
'gtf': '/data/dm6/r6-11/gtf/dm6_r6-11.gtf',
'chromsizes': '/data/dm6/r6-11/fasta/dm6_r6-11.chromsizes',
'bowtie2': '/data/dm6/r6-11/bowtie2/dm6_r6-11.1.bt2',
'hisat2': '/data/dm6/r6-11/hisat2/dm6_r6-11.1.ht2',
},
'r6-11_transcriptome': {
'fasta': '/data/dm6/r6-11_transcriptome/fasta/dm6_r6-11_transcriptome.fasta',
'chromsizes': '/data/dm6/r6-11_transcriptome/fasta/dm6_r6-11_transcriptome.chromsizes',
'salmon': '/data/dm6/r6-11_transcriptome/salmon/dm6_r6-11_transcriptome/hash.bin,
},
},
}